What is Informatics?
Protein modifications are often combinatorial and can occur at many sites within a protein, making data analysis and validation complex. Our In-house software is developed to meet these challenges, with the goal of developing tools and databases that can be a benefit throughout the scientific community.
Proteoform identification
The combinatorial nature of PTMs allows for hundreds or thousands of possible proteoforms per gene product that must be validated with data. ProSight is a software tool that allows the user to match mass spectra data with predicted proteoforms and their PTMs and provides a scoring metric to assess the quality of these matches. This software was developed in-house and is now commercialized. A free Windows application for matching mass spectral data against a single candidate protein sequence and its modifications is also available from the National Resource for Translational and Developmental Proteomics. (Proteaom. 2014, 15, 1235-1238.)
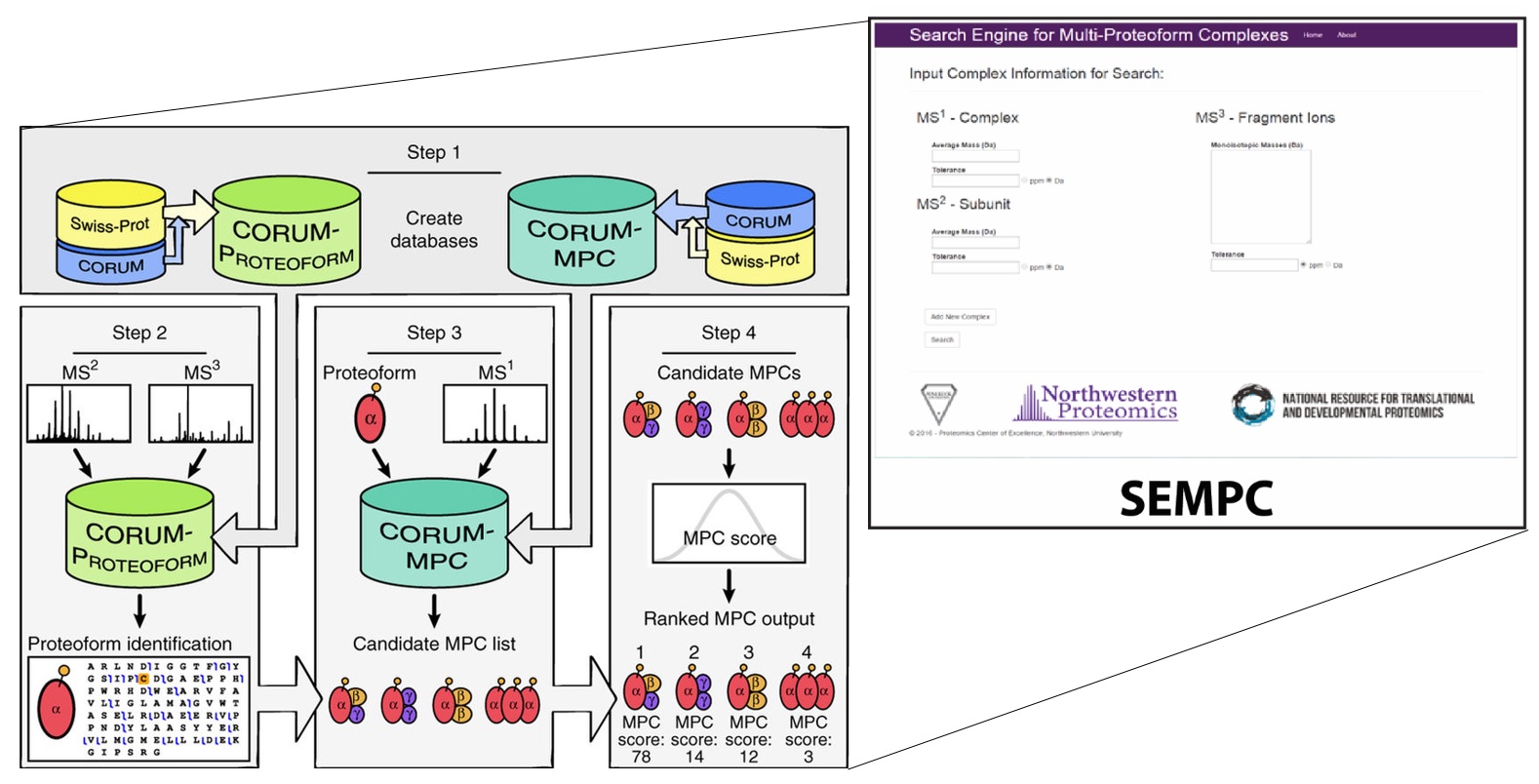
Structural characterization
Proteoforms often combine into non-covalent complexes. The possible combinations must be validated and scored as described above. SEMPC (“Search Engine for Multi-Proteoform Complexes”), is then used to characterize multimeric proteoform complexes. This search engine was also developed in-house. (Nat. Methods, 2016, 13, 237-240)
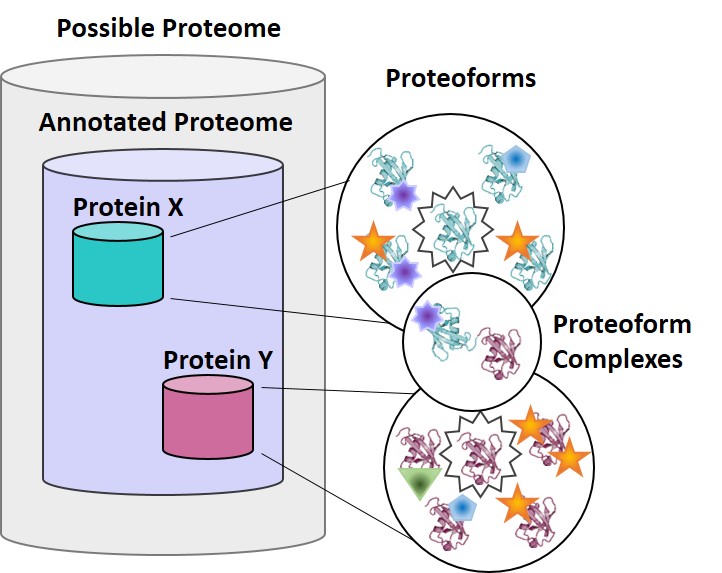
What is a proteoform?
A proteoform is the specific molecular form of a gene product, including any variation due to genetic mutation, alternative RNA splicing, and post-translational modifications (PTMs). Proteoforms have distinct biological functions, and are often difficult to characterize due to their subtle chemical differences. The Kelleher lab emphasizes innovation in proteomics tools and proteoform discovery.
