What is Natural Products Discovery?
Microbial natural products have exhibited great utility as drug-leads and molecular probes. Many of the most clinically important medicines, such as streptomycin (the first effective treatment for tuberculosis), doxorubicin (a potent cancer therapeutic), and penicillin (the first and one of the most widely used antibiotics) are produced by actinomycete bacteria or filamentous fungi. Historically, efforts to discover bioactive natural products from microbes has involved the painstaking process known as bioactivity-guided isolation. Despite early successes, recent decades using this approach have resulted in the frequent rediscovery of known natural products, giving the impression that researchers had exhausted nature’s reserve of new medicines. Fortunately, genomic studies--facilitated by a steady reduction in the cost of genome sequencing--have revealed that microbes possess far greater biosynthetic potential than previously realized and that natural product production genes are typically located together in biosynthetic gene clusters (BGC). Sequencing of actinomycetes and filamentous fungi has revealed that their genomes often contain dozens of BGCs responsible for the production of secondary metabolites. The disparity between the number of BGCs and the current rate of new natural product discovery justifies the need for novel discovery approaches.
Our natural products subgroup harnesses the power of high resolution mass spectrometry to identify and evaluate novel secondary metabolites from bacteria and fungi. We use both genomic and mass spectrometric techniques with sophisticated in-house bioinformatics tools for streamlined and incisive data analysis. Ultimately, we aim to generate a large library of novel natural product leads for early stage pharmaceutical discovery. Specific ongoing projects in the subgroup include: 1) Metabologenomics to identify novel metabolites with their biosynthetic gene clusters in actinomycete bacteria; 2) Fungal artificial chromosomes and metabolomic scoring (FAC-MS) to enable expression and identification of metabolites from otherwise cryptic gene clusters; 3) Chemical synthesis of novel natural products identified in the group; and 4) Biosynthetic investigation of novel natural products using platforms such as the phosphopantetheinyl ejection assay and Proteomic Investigation of Secondary Metabolism (PrISM). The natural products subgroup is highly interdisciplinary with collaborators in both academia and industry.
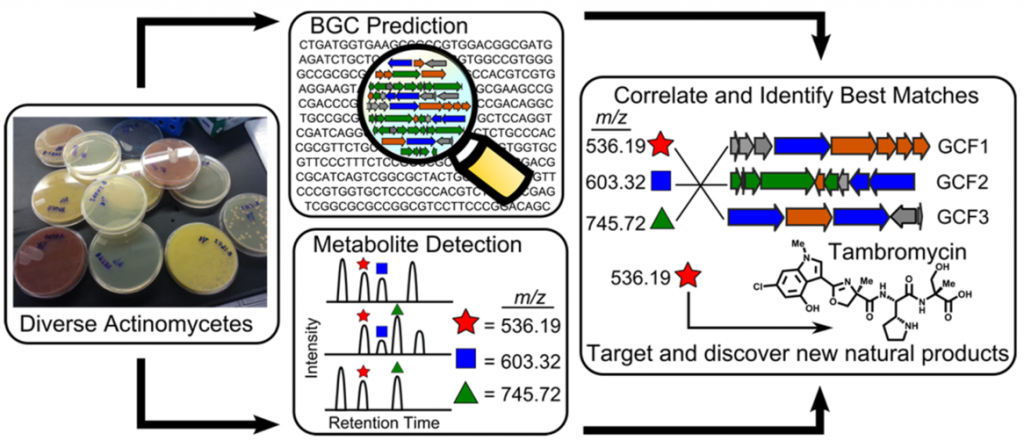
What is Metabologenomics?
As a collaboration with the Metcalf lab at the University of Illinois at Champaign-Urbana and the Thomson lab at Northwestern, we have assembled biosynthetic gene cluster families (GCFs) from hundreds of sequenced and annotated actinobacterial genomes. These data have been combined with high-accuracy mass spectrometric (LC-MS) analysis of extracts generated from the same actinobacteria to generate a simple correlation score that rates the association of an observed natural product with a GCF based upon their co-occurrence pattern across the strain library. This method of combining metabolomics and genomics to simultaneously illuminate novel chemical structures with their biosynthesis at large scale is known as metabologenomics. The metabologenomics platform provides a new screen for secondary metabolites whose data can be mined to identify new members of interesting structural classes, to determine the biosynthetic origin of a known natural product, or to identify entirely new structural classes and biosyntheses.

FAC-MS (Fungal Artificial Chromosomes and Metabolomic Scoring)
The genomes of filamentous fungi contain between 50-80 biosynthetic gene clusters, and thus represent an untapped wealth of novel natural products. Because most of these gene clusters are transcriptionally “silent” under normal laboratory conditions, the vast majority of these metabolites are inaccessible to traditional natural product discovery approaches. In collaboration with the Keller lab at UW-Madison and industry partner Intact Genomics we have a developed FAC-MS, a fungal natural products discovery platform that bypasses this problem through heterologous expression of intact gene clusters in the model fungus, Aspergillus nidulans, and identification of novel metabolites via mass spectrometry-based metabolomics. We recently used this platform for discovery of a novel lipopeptide macrolactam, valactamide A, as well as the biosynthetic pathway for the orphan benzodiazepine, benzomalvin A/D.
