What is Native Proteomics?
Most proteins must non-covalently interact with other partners to be biologically active; these protein complexes are critical for efficient catalysis and cellular signaling. “Native” top-down mass spectrometry can be used to characterize non-covalent interactions including protein-protein and protein-cofactor binding.
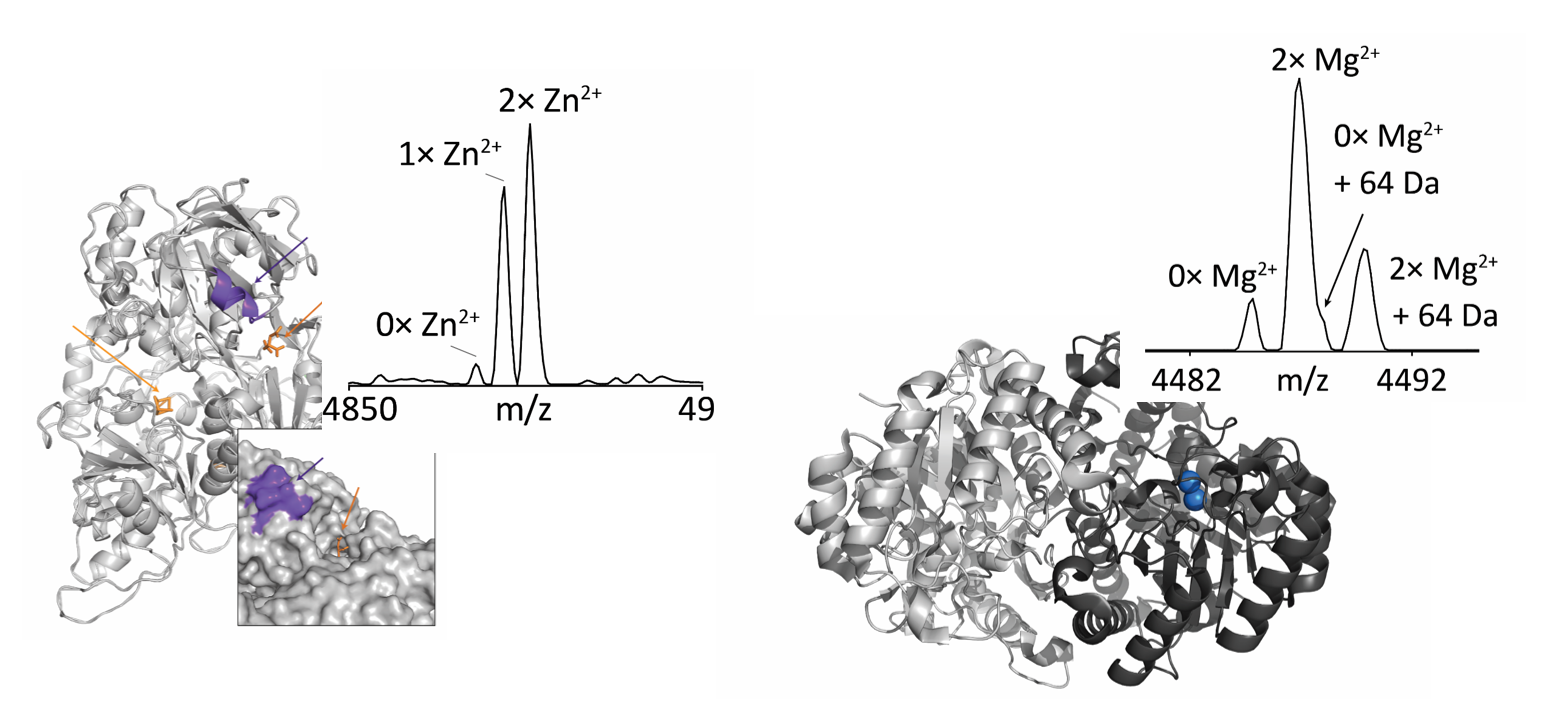
Metal binding in metalloenzymes
Native top-down is the analysis of folded, intact proteins. This technique provides a direct readout of protein complexes and their non-covalent binding partners, such as metal cofactors. Shown are intact mass spectra of aconitase (left) and enolase (right). Nat. Chem. Biol. 2017, 14, 36-41
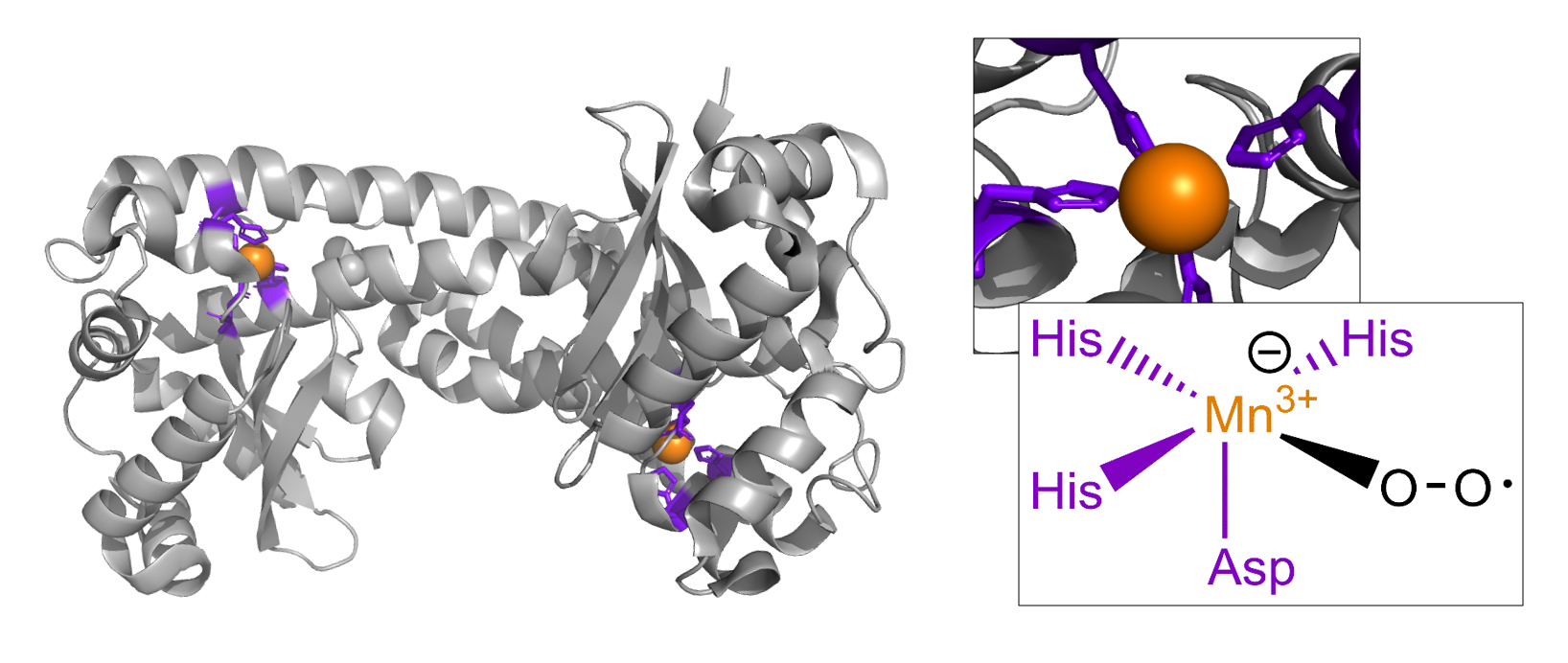
Ternary complexes
Native top-down can be used to characterize labile substrates bound to protein complexes. Shown are intact mass spectra of superoxide intermediates bound to mitochondrial superoxide dismutase. Nat. Chem. Biol. 2017, 14, 36-41
What is a proteoform?
A proteoform is the specific molecular form of a gene product, including any variation due to genetic mutation, alternative RNA splicing, and post-translational modifications (PTMs). Proteoforms have distinct biological functions, and are often difficult to characterize due to their subtle chemical differences. The Kelleher lab emphasizes innovation in proteomics tools and proteoform discovery.
